使用 MSMBuilder 进行数据聚类
之前的一篇博文讲述了通过scipy.cluster进行聚类,最近发现MSMBuilder也具有类似的功能,并且使用比较方便,遂记录于此。
准备工作
在构建 MSM 的过程中,一般会有如下降维的步骤: 1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18from msmbuilder.featurizer import RawPositionsFeaturizer
from msmbuilder.decomposition import PCA
from msmbuilder.cluster import MiniBatchKMeans
featurizer = RawPositionsFeaturizer(atom_indices=atoms)
feat = featurizer.fit_transform(trajs) # trajs 为列表,每个元素为一条轨迹;feat 为列表,每个元素为 帧数* 特征数 的 2D array
pca = PCA(n_components=2)
pca_traj = pca.fit_transform(feat) # pca_traj 为列表,长度为轨迹条数;每个元素为 帧数 * n_components 的 2D array
pca_data=np.concatenate(pca_traj)
# 开始聚类
n_clusters=200
clusterer = MiniBatchKMeans(n_clusters=n_clusters, random_state=1)
clustered_traj = clusterer.fit(pca_traj) # pca_traj 为列表
clusterer.labels_ # 聚类标签
clusterer.cluster_centers_ # 聚类中心 RawPositionsFeaturizer提取特征,然后使用PCA进行降维,再使用MiniBatchKMeans进行聚类。需要注意的是:
1. clusterer.fit(pca_traj) 表示使用pca_traj来
fit 模型的参数 2. clusterer.transform(new_data) 表示计算
new_data 在这个模型上的投影 3. pca_traj
是一个列表,它的每一个元素是一组数据 4.
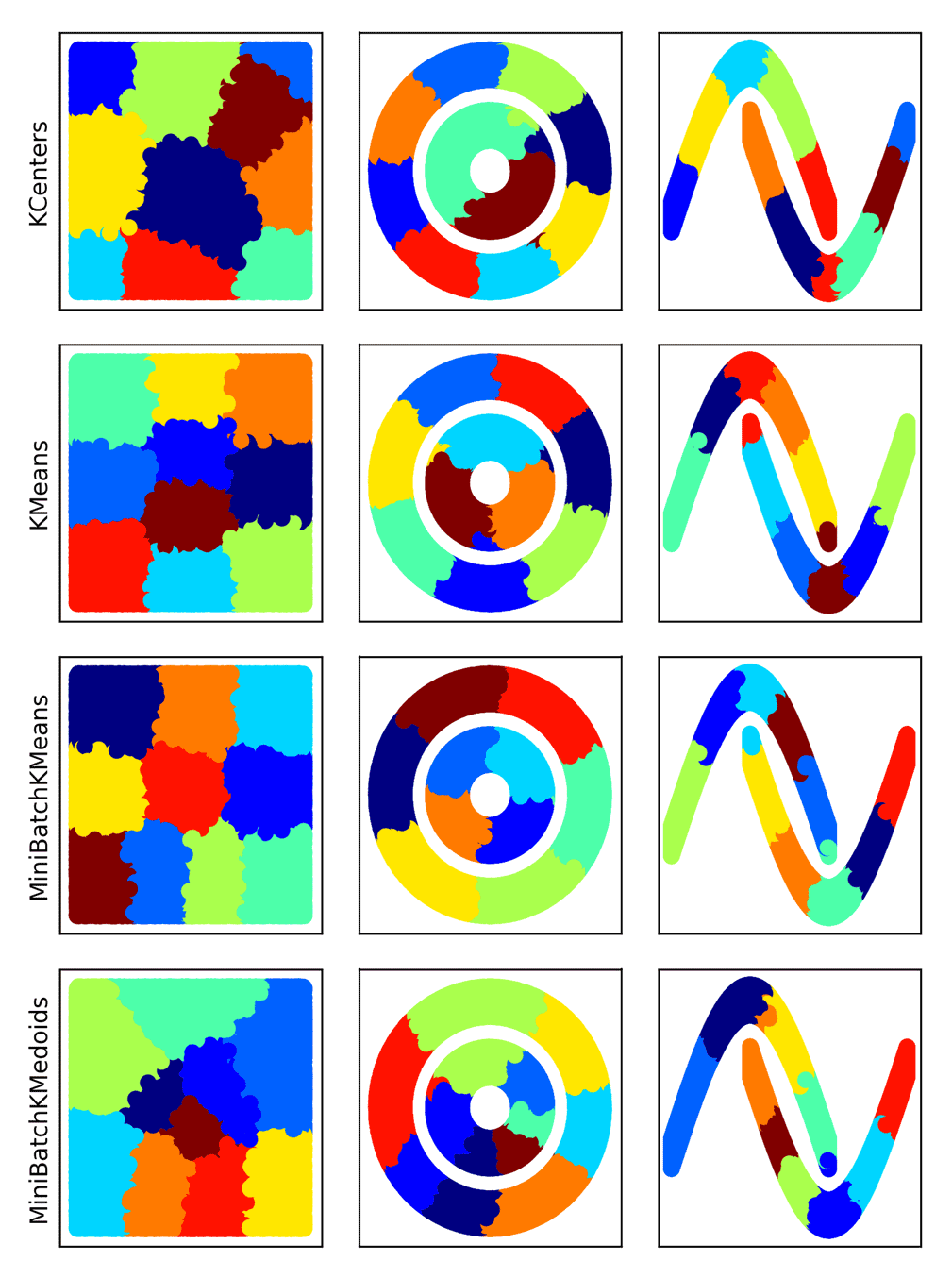
除了MiniBatchKMeans之外,msmbuilder
还提供了其它的聚类方案:
| API | Description |
|---|---|
| KCenters | K-Centers clustering |
| KMeans | K-Means clustering |
| KMedoids | K-Medoids clustering |
| MiniBatchKMedoids | Mini-Batch K-Medoids clustering |
| RegularSpatial | Regular spatial clustering |
| LandmarkAgglomerative | Landmark-based agglomerative hierarchical clustering |
| AffinityPropagation | Perform Affinity Propagation Clustering of data |
| GMM | Gaussian Mixture |
| MeanShift | Mean shift clustering using a flat kernel |
| MiniBatchKMeans | Mini-Batch K-Means clustering |
| SpectralClustering | Apply clustering to a projection to the normalized laplacian |
| Ward |
测试
1 | %pylab |
效果如下: